3DBrainMiner is a analysis framework developped in collaboration between the LISTIC (USMB), the LIFAT (U.Ttours) and the NECOS team of INRAE Nouzilly.
This research aims at developing models and algorithms to describe, analyze and compare objectively the brains of different animal species or humans.- These current works aim at:
- i/ making images, segmentations, and graph representations of brain available to the Neuroscience, Animal Sciences and Machine Learning research communities =>3DBrainModels
- ii/ providing open access to the
source codes of the tools developped to generate graph
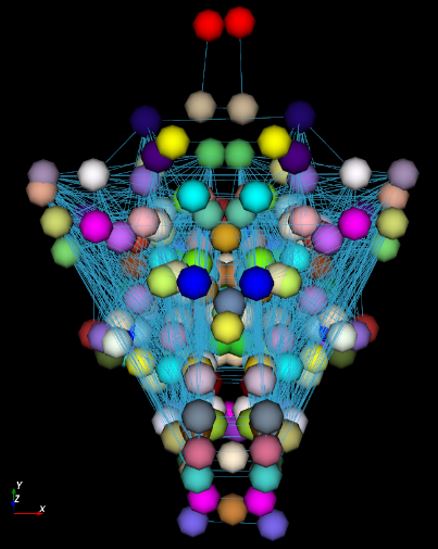
representations of the brains => 3DBrainMiner
platform.
The produced graphs can combine a wide variety of information such as morphometric (volume, distance, ...), structural (signal intensity) and functional (symmetry, connection strength, ...) variables. - iiI/ designing new algorithms dedicated to the visualisation, the analysis and comparison of graphs (using Graph Neural Networks)
- In the context of Antoine Bourlier's PhD thesis (co-direction LISTIC, LIFAT, NECOS, co-funding INRAE-Région CVL), Graph-based machine learning algorithms (GNN) are developed to analyze and compare the models produced with their various parameters to, for example, assess the impact of the rearing environment (type of suckling, ...) on the anatomical organization of the brain of young lambs.
- This dynamic will allow a better understanding of the anatomy of the ovine brain, its development and the impact of the rearing conditions. In the longer term, this platform could be extended to other animal species to facilitate comparative anatomy and to human brains for clinical purposes.
_
LLISTIC - USMB. - LIFAT - Université de Tours.
NECOS -INRAE Tours
Contacts: Jean-Yves Ramel, Elodie Chailliou